
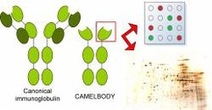
Unlike Laboratory settings, in which growth conditions can be controlled and changed one at a time, bacteria in the Environment must perpetually make transcriptional decisions between activating metabolic genes for available, frequently mixed C-sources and those for escaping or adapting to physico-chemical stress. Our research is committed to the development of novel strategies for construction of soil microorganisms (mostly Pseudomonads) destined for the environment or as catalysis for selected biotransformations. To this end we employ Systems and Synthetic Biology as a source of new tools for addresing some outstanding environmental pollution problems.

Cells exhibit a wide range of dynamical behaviours which allow them to interpret their environment and to function as social organisms. We are interested in understanding these behaviours by combining two approaches. First, we study how genetic circuits regulate function in a mechanistic way. To this aim, we apply quantitative modelling and experiments in both natural and synthetic systems. We then complement this approach with evolutionary studies in order to understand why particular circuit functions and structures have been selected. This we extend to the analysis of the structure of the genome, the material where genetic circuits and networks are encoded

The biological functions of many proteins can only be explained in the context of their relationshipts with others. The study of living systems from a network perspective is providing new biological knowledge which could have never been obtained from the study of the individual components (genes, proteins, ...) no matter how detailed it is. Our group is interesting in finding this biological knowledge "hidden" in the complex network of relationships between biological elements. We study biological networks (specially protein interactions networks and metabolic networks) from this approach. We are also very interested in approaching the complex phenomenon of "protein function" from a systemic perspective.
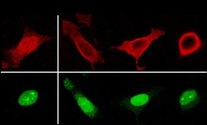
Integration of external stimuli and initiation of the appropriate genetic response are key events in the function of the nervous system. Our work is focused on two major signaling pathways, calcium and cAMP, and two main transcriptional master-switch regulators, CREB and DREAM, that convey the activity-dependent response from single cells to neuronal circuits. Using transcriptomic and proteomic approaches we aim to define gene populations and protein-protein interactions defining transcriptional networks that specifically shape each genetic response.