Golden Standard Tutorial
GS Recovery from the filter paper
GS Wizard overview:
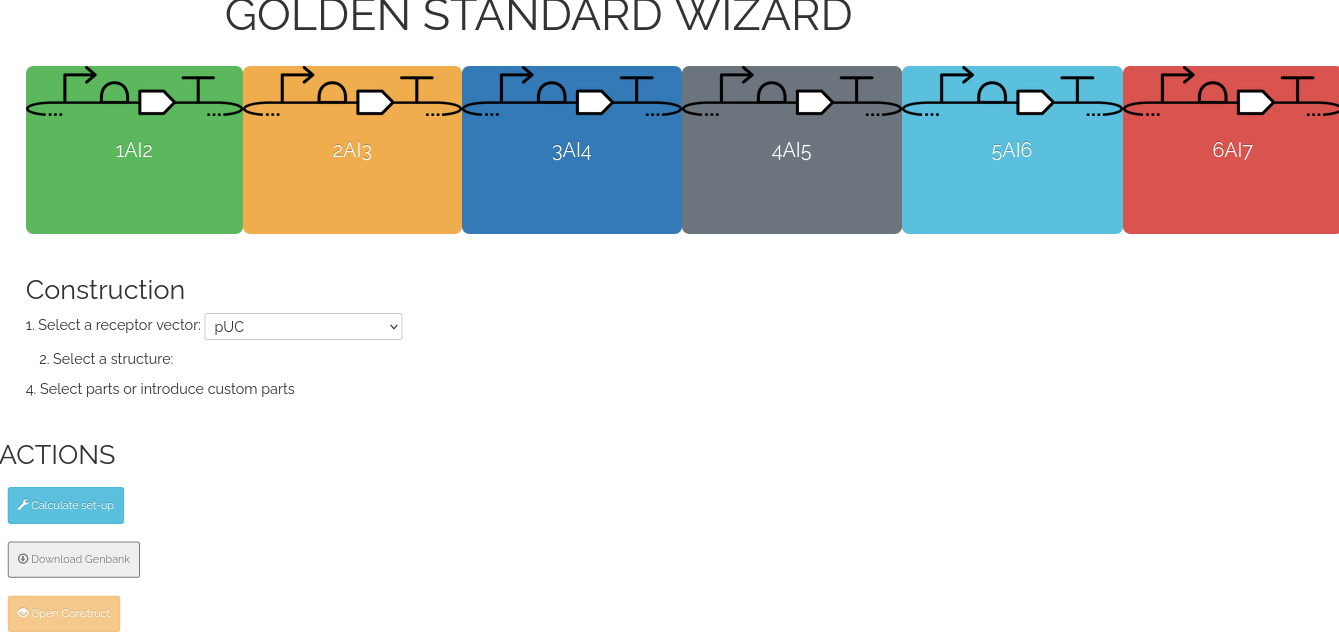
The tool is dividied in multiple sections. The upper part contains the TU to be selected by the user, clicking the color squares.
The middle are is the blackboard where it is possible to select the receptor vector, the structure and the parts to create a construction. And the bootom part is the section with the multiple actions to do with the construction.
Note: This wizard allows the assembly of maximun of 6 TUs, the process to create a more complex assembly can be found in (Blazquez et al 2022)
Procedure
Step 1. Select a receptor vector
Golden Standard contains three possibilities, PUC, RK2 y pBRR1

Step 2. Structure selection. With zero, one or multiple tags.
The user can select the option to design a basic structure with promoter, RBS, cds and terminator or include tags in N-terminal or C-terminal position.

Based on the structure selection, the construction showed in the bottom panel is changing to acomodate the new parts.

2.1 Design
The first step is the TU selection clicking on the colored squares in the top section.

2.1.1 Custom level 1 constructions.
2.2.1 Select Golden Standard parts
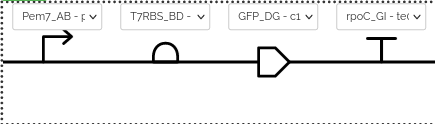
- Click on a specific part (squared name) and select a element. It can be repeated as many times as elements to edit. The following image shows the output of the process after selection of the promoter Pem7, the RBS T7RBS, and so on. It is important to select the right parts based on their fusion sites, for example, if N-Tag is included, only a RBS with a BC fusion site is right. The tool shows all of them but only a RBS with a BC fusion site can give a right construction.
2.1.2 Select non Golden Gate parts
Previous requirements:
When a new sequence is included into the server some previous steps are required to perform a correct assembly.
New Sequence protocol:
1. If the assembly is basic (Promoter+RBS+CDS+Terminator):
-
Part
Position
modifications
Fixed sequences
Manual check point
Promoter
AB
Remove restriction enzymes
5'
GGTCTCAGGAG
3'
TACTAGAGACC
RBS
BD
Remove restriction enzymes
5'
GGTCTCATACT
3'
AATGAGAGACC
CDS
DG
Remove restriction enzymes
5'
GGTCTCAAATG
stop codon included
Remove ATG from sequence
3'
GCTTAGAGACC
Terminator
GI
Remove restriction enzymes
5'
GGTCTCAGCTT
3'
CGCTAGAGACC
2. If the assembly contains tags:
2.1 One N-tag:
The sequence require the following modifications, change the sequence according the type of the part.
-
Number of tags Part type Position modifications orientation Fixed sequences Manual check point 1 x N-Tag
Promoter
AB
5'
GGTCTCAGGAG
3'
TACTAGAGACC
RBS
BC
5'
GGTCTCATACT
Distance to ATG
3'
CCATAGAGACC
N-tag_1
CD
Remove ATG from original sequence
5'
GGTCTCACCATg
Reading frame
3'
gcAATGAGAGACC
CDS
DG
Remove ATG from original sequence
5'
GGTCTCAAATG
Reading frame
3'
GCTTAGAGACC
stop codon included
Terminator
GI
5'
GGTCTCAGGCT
3'
CGCTAGAGACC
2x N-tags
Promoter
AB
5'
GGTCTCAGGAG
3'
TACTAGAGACC
RBS
BC
5'
GGTCTCATACT
Distance to ATG
3'
CCATAGAGACC
N-tag_1
CD
Remove ATG from original sequence
5'
GGTCTCACCATg
Reading frame
3'
gcAATGAGAGACC
N-tag_2
DE
Remove ATG from original sequence
5'
GGTCTCAAATG
Reading frame
3'
tcAGGTAGAGACC
CDS
EG
Remove ATG from original sequence
5'
GGTCTCAAGGTcg
stop codon included
3'
GCTTAGAGACC
Terminator
GI
5'
GGTCTCAGCTT
3'
CGCTAGAGACC
1 x C-Tag
Promoter
AB
Remove ATG from original sequence
5'
GGTCTCAGGAG
3'
TACTAGAGACC
RBS
BD
5'
GGTCTCATACT
Distance to ATG
3'
AATGAGAGACC
CDS
DF
Remove ATG from original sequence
5'
GGTCTCAAATG
3'
TTCGAGAGACC
No stop codon
C-tag_1
FG
5'
GGTCTCACCATgc
Reading frame
3'
GCTTAGAGACC
Stop codon included
Terminator
GI
5'
GGTCTCAGCTT
3'
CGCTAGAGACC
2x C-tags
Promoter
AB
5'
GGTCTCAGGAG
3'
TACTAGAGACC
Distance to ATGHola
RBS
BD
5'
GGTCTCATACT
3'
AATGAGAGACC
CDS
DF
Remove restriction enzymes
5'
GGTCTCAAATG
Remove ATG from sequence
3'
TTCGAGAGACC
C-tag_1
FG
Remove restriction enzymes
5'
GGTCTCATTCGgc
Reading frame
Remove ATG from sequence
3'
GCTTAGAGACC
C-tag_2
GH
Remove restriction enzymes
5'
GGTCTCATTCGcg
Reading frame
Remove ATG from sequence
3'
GGTAAGAGACC
Terminator
HI
Remove restriction enzymes
5'
GGTCTCAGGTA
3'
CGCTAGAGACC
1 x N-Tag + 1x C-Tag
Promoter
AB
Remove restriction enzymes
5'
GGTCTCAGGAG
3'
TACTAGAGACC
RBS
BC
Remove restriction enzymes
5'
GGTCTCATACT
Distance to ATG
3'
CCATAGAGACC
N-tag_1
CD
Remove restriction enzymes
5'
GGTCTCACCATg
Reading frame
Remove ATG from sequence
3'
gcAATGAGAGACC
CDS
DF
Remove restriction enzymes
5'
GGTCTCAAATG
Remove ATG from sequence
3'
TTCGAGAGACC
Remove stop codon
C-tag_1
FG
Remove restriction enzymes
5'
GGTCTCATTCGgc
Reading frame
Remove ATG from sequence
3'
GCTTAGAGACC
Terminator
GI
Remove restriction enzymes
5'
GGTCTCAGCTT
3'
CGCTAGAGACC
2x N-tags + 1 C-tag
Promoter
AB
Remove restriction enzymes
5'
GGTCTCAGGAG
3'
TACTAGAGACC
RBS
BC
Remove restriction enzymes
5'
GGTCTCATACT
Distance to ATG
3'
CCATAGAGACC
N-tag_1
CD
Remove restriction enzymes
5'
GGTCTCACCATg
Reading frame
Remove ATG from sequence
3'
gcAATGAGAGACC
N-tag_2
DE
Remove restriction enzymes
5'
GGTCTCAAATG
Reading frame
Remove ATG from sequence
3'
tcAGGTAGAGACC
CDS
EF
Remove restriction enzymes
5'
GGTCTCAAGGTcg
Remove ATG from sequence
3'
TTCGAGAGACC
C-tag_1
FG
Remove restriction enzymes
5'
GGTCTCATTCGgc
Reading frame
Remove ATG from sequence
3'
GCTTAGAGACC
Terminator
GI
Remove restriction enzymes
5'
GGTCTCAGCTT
3'
CGCTAGAGACC
1 x N-Tag + 2 x C-Tag
Promoter
AB
Remove restriction enzymes
5'
GGTCTCAGGAG
3'
TACTAGAGACC
RBS
BC
Remove restriction enzymes
5'
GGTCTCATACT
Distance to ATG
3'
CCATAGAGACC
N-tag_1
CD
Remove restriction enzymes
5'
GGTCTCACCATg
Reading frame
Remove ATG from sequence
3'
gcAATGAGAGACC
CDS
DF
Remove restriction enzymes
5'
GGTCTCAAATG
Remove ATG from sequence
3'
TTCGAGAGACC
Remove stop codon
C-tag_1
FG
Remove restriction enzymes
5'
GGTCTCATTCGgc
Reading frame
Remove ATG from sequence
3'
GCTTAGAGACC
C-tag_2
GH
Remove restriction enzymes
5'
GGTCTCAGCTTcg
Reading frame
Remove ATG from sequence
3'
GGTAAGAGACC
Terminator
HI
Remove restriction enzymes
5'
GGTCTCAGGTA
3'
CGCTAGAGACC
2 x N-Tag + 2 x C-Tag
Promoter
AB
Remove restriction enzymes
5'
GGTCTCAGGAG
3'
TACTAGAGACC
RBS
BC
Remove restriction enzymes
5'
GGTCTCATACT
Distance to ATG
3'
CCATAGAGACC
N-tag_1
CD
Remove restriction enzymes
5'
GGTCTCACCATg
Reading frame
Remove ATG from sequence
3'
gcAATGAGAGACC
N-tag_2
DE
Remove restriction enzymes
5'
GGTCTCAAATG
Reading frame
Remove ATG from sequence
3'
tcAGGTAGAGACC
CDS
EF
Remove restriction enzymes
5'
GGTCTCAAGGTcg
Remove ATG from sequence
3'
TTCGAGAGACC
Remove stop codon
C-tag_1
FG
Remove restriction enzymes
5'
GGTCTCATTCGgc
Reading frame
Remove ATG from sequence
3'
GCTTAGAGACC
C-tag_2
GH
Remove restriction enzymes
5'
GGTCTCAGCTTcg
Reading frame
Remove ATG from sequence
3'
GGTAAGAGACC
Terminator
HI
Remove restriction enzymes
5'
GGTCTCAGGTA
3'
CGCTAGAGACC
Once, the sequence is corrected, it can be included in the system.
Click the combo-box and select “Custom part”. A pop up window is opened and it can be fill with the custom name, the sequence or the size of the sequence and the concentration. The following figure shows the case where the cds is selected.

b) Click on Submit button to save the changes.
Step 3. Set up calculations
After the selection of all parts, the final information related to final concentration, enzyme requirements, required water and buffer quantity can be calculated clicking on the button “Calculate set-up“
![]()
Step 4. See the results
The construction can be stored as genbank format clicking on the Download Genbank Button, or can be viewed by the viewer tool or it can be set up.

Step 4.1 Setup
The concentrations and sequence lengths form the assembly are used to calculate the final protocol parameters. The Golden Gate Wizard tool send the information to Golden Gate Setup module and redirect to Golden Standard Setup page.

A table with the information introduced previously appear, with the final calculations. The results can be easily edited clicking on the squares. Initially, the concentrations are empty and it is necessary to fill them to have final concentrations instead infinity values. In the upper part of the table the total amount of volume or the number of desired fmol can be modified if the user needs.

After the calculation is done, this table can be printed using the button Print.
Step 4.2. View the construction
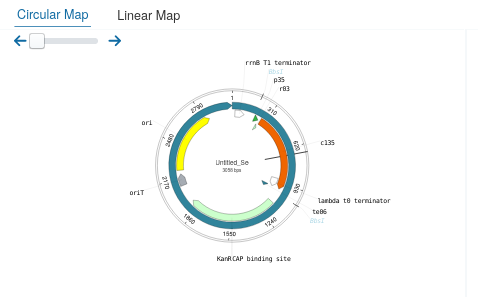
To store the construction, it can be downloaded clicking the Export button.
The server will redirect to the vector viewer (Open-vector-editor)
showing the final construction. By default Single cutters restriction sites are activated, but to show the restriction sites of the enzymes used in Golden Standard it can be activated Clicking on .
A menu will open with the activated enzymes. And to include others, clicking on the top-right triangle a list of them will be shown.
GoldenStandard can be selected and it will be included into the activated enzymes.
Step 4.3. Download the construction
To download the construction in Genbank format, it is necessary click on Download genbank button.
